| Browse |
|
2.1 Peptides
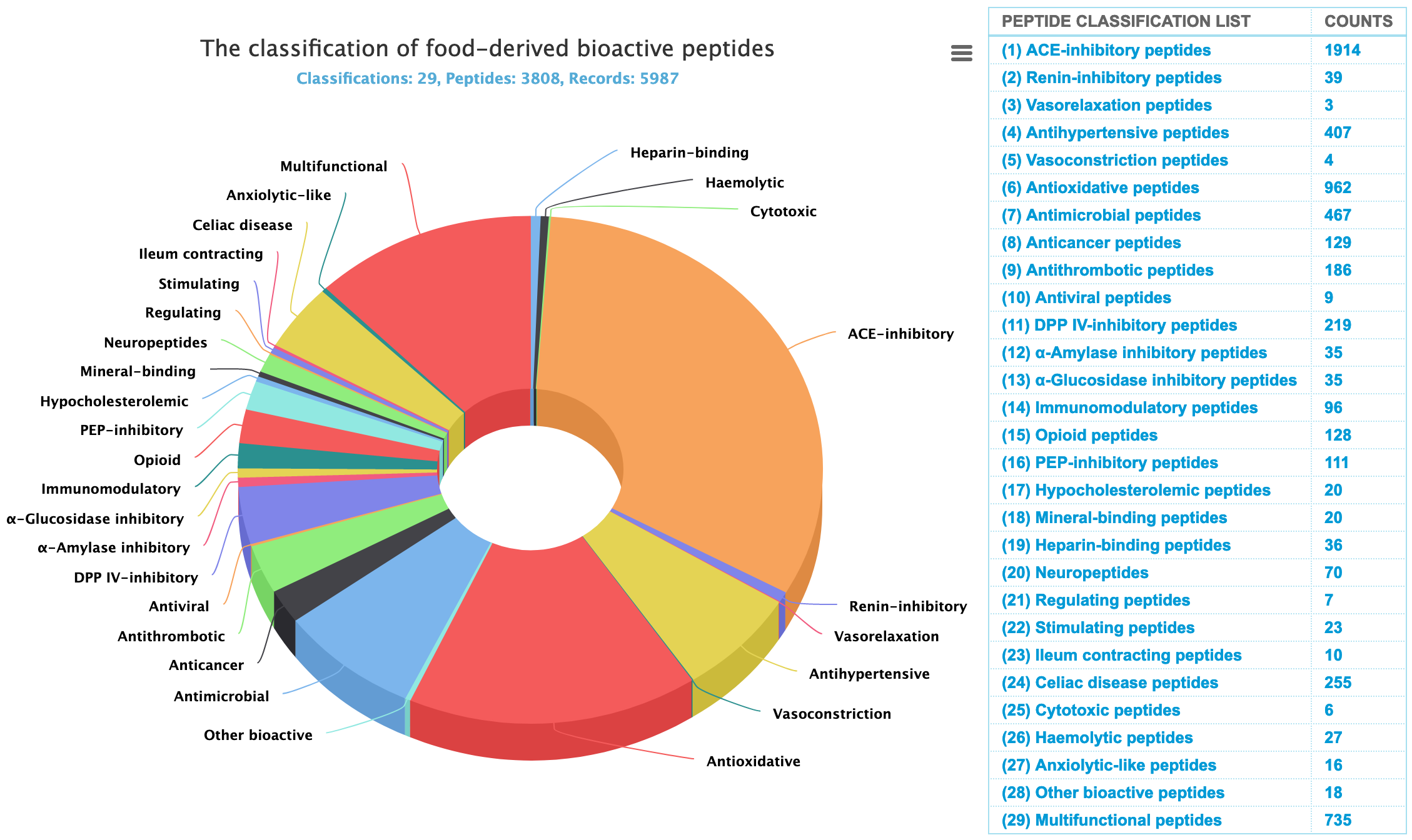
2.1.1 Peptide classification DFBP currently contains 5987 peptide entries among which 3808 are unique peptides. These peptides are divided into 29 types according to their functions, particularly including 745 multifunctional peptides.
|
- PathWay Map: KEGG reported bioactive peptide target protein metabolism pathway (https://www.genome.jp/kegg/pathway.html).
- MultifunPep Search: To find the relationship between the current bioactive peptides and other 28 kinds of bioactive peptides. Through search and analysis, you can find multifunctional peptides and study the mechanism connection between different bioactive peptides.
- Statistical data: To perform statistical analysis of attribute information of bioactive peptides, including C-terminal and N-terminal amino acid types, peptide length distribution, peptide amino acid composition, and relative molecular mass distribution, etc.
- Advanced Search: To turn to the advanced search page for detailed filtering operations.
|
- Search: To quickly search for bioactive peptides by DFBP ID, AA sequence, Peptide name, AA length, Precursor protein and PubDate. In order to ensure the convenience of searching, the letter size is not distinguished here.
- Display: The number of peptides displayed on each page can be adjusted by "Display". "First, previous, next, last" is used to quickly turn pages. You can also use "Page to" to directly turn to the specific page.
|
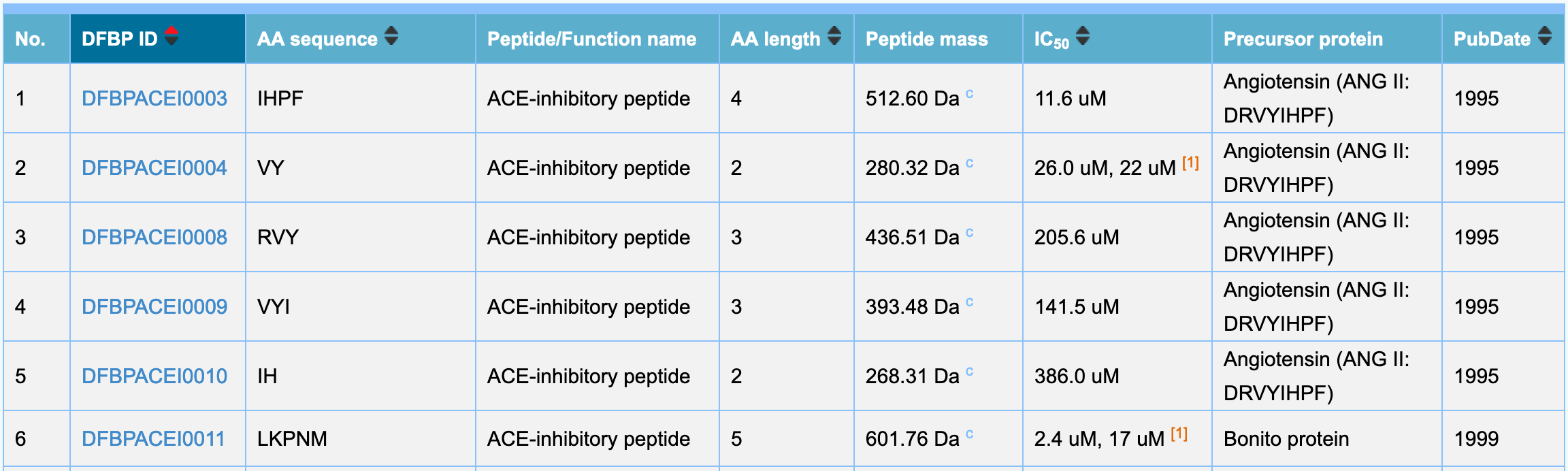
- The brief list is used to display basic information of bioactive peptides, including DFBP ID, AA sequence, Peptide/Function name, AA Length, Peptide mass, IC50, Precursor protein and PubDate.
- To quickly filter by entering the corresponding information in the quick search tool, results matching the search criteria will be highlighted in the list. If you need more advanced filtering operations, please click "Advanced Search".
- To provide sorting operations for related attributes, including DFBP ID, AA sequence, AA Length, IC50 and PubDate.
- If you need detailed information for a peptide, please click "DFBP ID" number to enter the peptide attribute page.
|
2.1.3 Peptide property page This page details 30 kinds of attributes of each peptide, mainly divided into the following 12 parts:
(1) Peptide header information
- DFBP ID: The unique ID identification number assigned by DFBP.
- Peptide sequence: Single letter amino acid sequence of a peptide.
- Type: The peptides included in DFBP are basically natural food-derived bioactive peptides, thus they are all considered to be native peptides.
- Peptide name: Named according to the function of a peptide, or defined in the literatures.
|
(2) Functional-activity relationship
- Main bioactivity: The bioactivity of the peptide reported in the literature.
- Other bioactivity: In addition to the bioactivity according to the current classification, whether it has other bioactivities. In this example, in addition to ACE-inhibitory activity, it also exhibit Antioxidative, Antioxidative, DPP-IV inhibitory activity. There are total four bioactivities, so this peptide belongs to a multifunctional bioactive peptide.
|
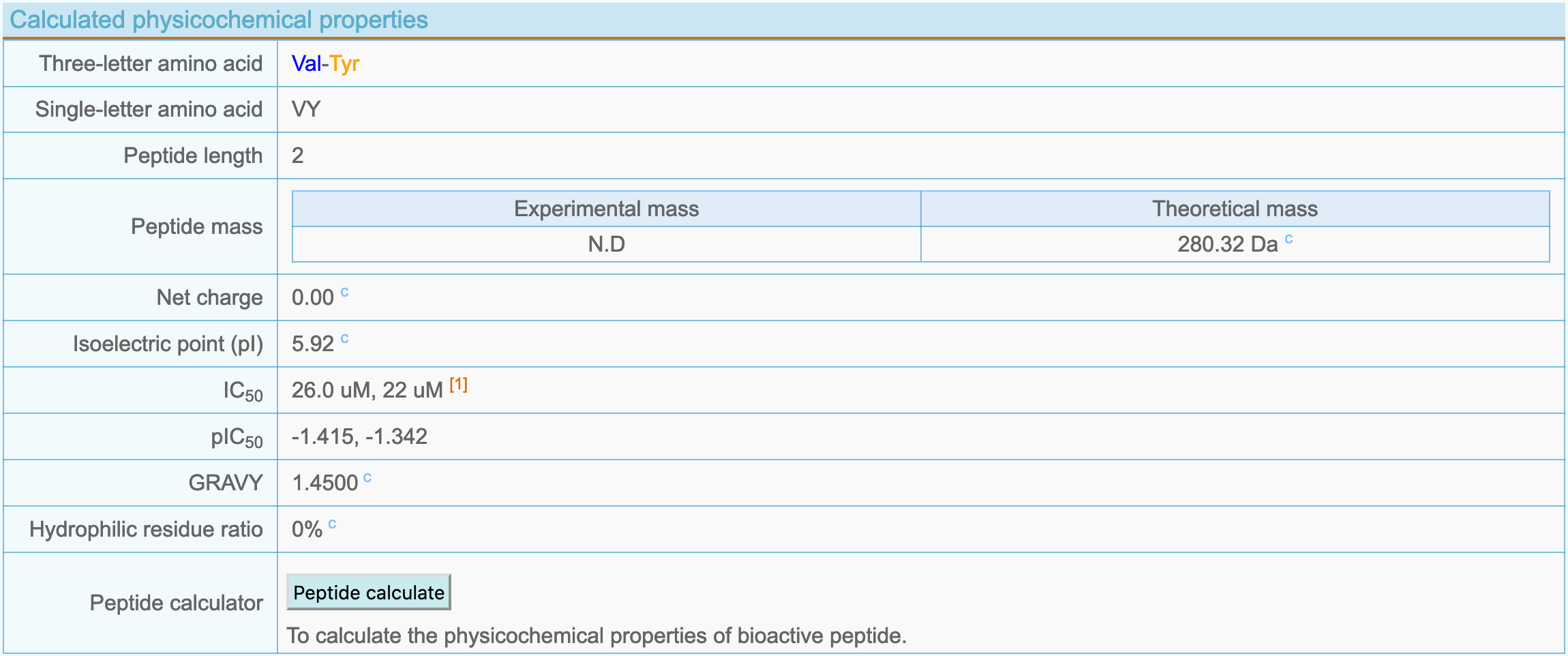
(3) Calculated physicochemical properties
- Three-letter amino acid: Peptide 3-letter amino acid sequence.
- Single-letter amino acid: Single letter amino acid sequence of a peptide.
- Peptide length: The number of amino acid residues.
- Peptide mass: The actually measured relative molecular mass of the peptide reported in the article and the calculated theoretical mass.
- Net charge: Peptide net charge (pH = 7.0).
- Isoelectric point (pI): Peptide isoelectric point.
- IC50: The IC50 value of the bioactive peptide reported in the article.
- pIC50: pIC50 = -logIC50.
- GRAVY: Grand average of hydropathicity.
- Hydrophilic residue ratio: Proportion of hydrophobic amino acid residues in a peptide.
- Peptide calculator: Three types of calculation tools are provided, all of which calculate the relevant physicochemical properties based on the peptide sequence. The "Peptide property calculator" is a calculation tool provided by DFBP, and the other two are provided by ExPASy and NovoPro.
|
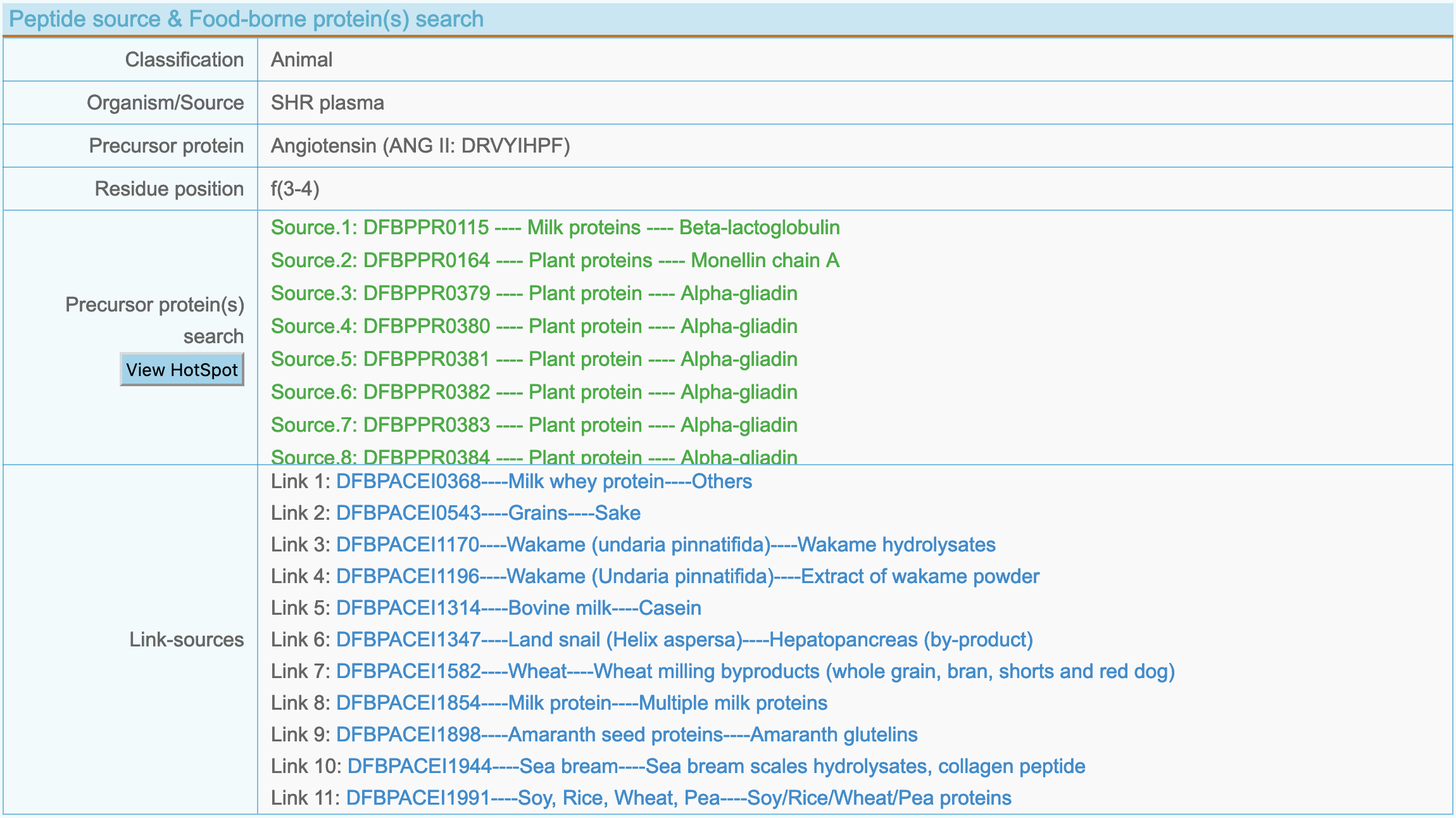
(4) Peptide source and food-borne protein(s) search
- Classification: Food-derived bioactive peptides are divided into 5 categories: Milk, Plant, Animal, Marine and Microorganism.
- Organism(s): The organism of the peptide reported in the article.
- Precurso protein: Precursor protein of bioactive peptides.
- Residue position: Distribution site of peptide in the precursor protein.
- Precursor protein(s) search: Food-borne precursor proteins list containing bioactive peptide fragments. To view the distribution of the peptide in food-borne proteins by click "View HotSpot".
- Link-sources: In addition to the current article reports, other articles that have studied the same sequence. To view specific information by click a link.
|
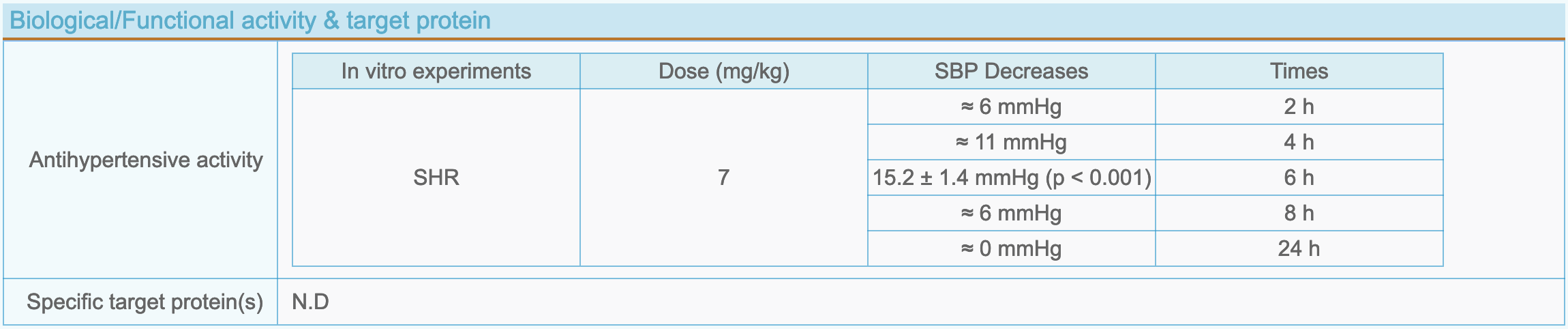
(5) Peptide biological activity and target protein
- Antihypertensive activity: The bioactivity data reported in the literatures, including detailed experimental data, graphical information and evaluation of experimental results, etc.
- Specific target protein(s): Target protein interacting with the peptide.
|
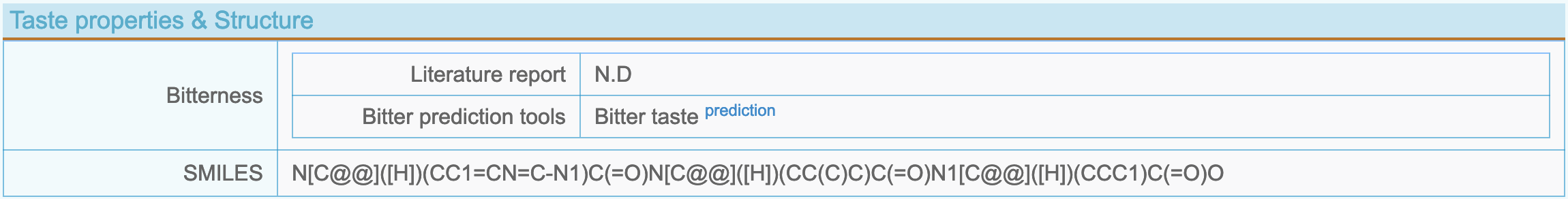
(6) Peptide taste and structure
- Bitterness: Bitterness information of a peptide, including literature report, database retrieval and bitterness prediction.
- SMILES: Generated by PepSMI (https://www.novoprolabs.com/tools/convert-peptide-to-smiles-string).
|
(7) Peptide preparation method
- Mode of preparation: Preparations of peptide reported in the literature, there are three main methods: enzymatic hydrolysis, peptide synthesis and microbial fermentation.
- Enzyme(s)/starter culture: Enzymes, synthesis method and fermentation conditions for preparing bioactive peptides.
|
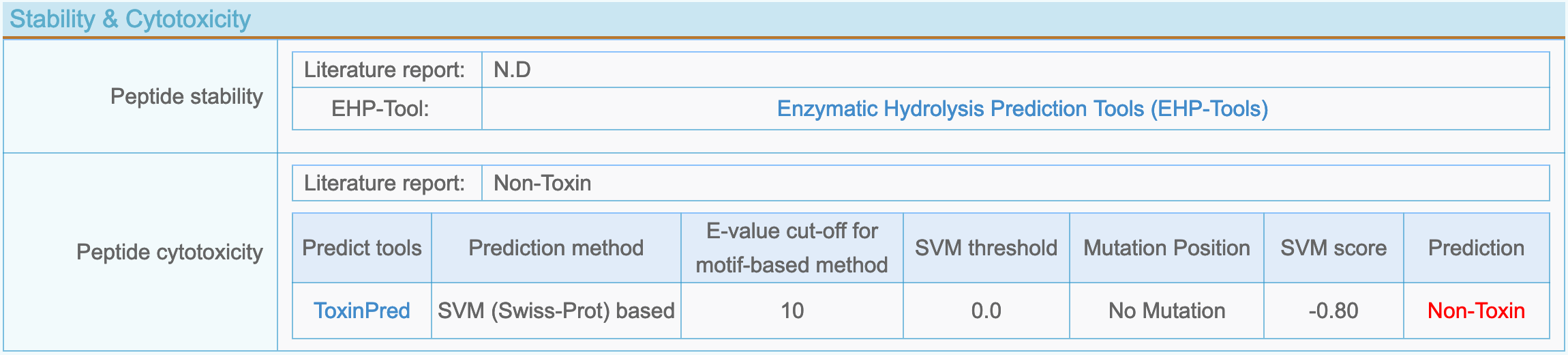
(8) Peptide stability and cytotoxicity
- Peptide stability: Thermal stability, pH stability, enzyme stability reported in the literature and two enzyme hydrolysis prediction tools: EHP-Tool (DFBP), Peptidecutter (https://web.expasy.org/peptide_cutter/).
- Peptide cytotoxicity: Peptide toxicity reported in the literature and toxicity predicted by ToxinPred (http://crdd.osdd.net/raghava/toxinpred/design.php).
|
(9) Additional information
- Additional information: Supplementary information about other relevant data of a peptide.
|
(10) Database cross-references
- DFBP: Multifunctional bioactivities links in DFBP database.
- BIOPEP: Link to BIOPEP database(http://www.uwm.edu.pl/biochemia/index.php/en/biopep).
- APD: Link to APD database(http://aps.unmc.edu/AP/database/query_input.php).
- BioPepDB:Link to BioPepDB database(http://bis.zju.edu.cn/biopepdbr/index.php?p=search).
- MBPDB: Link to MBPDB database(http://mbpdb.nws.oregonstate.edu/peptide_search/).
|
- Primary literature: The article that mainly reports about the peptide.
- Other literature(s): Other articles related to the peptide.
- Pubdate: Publication date.
|
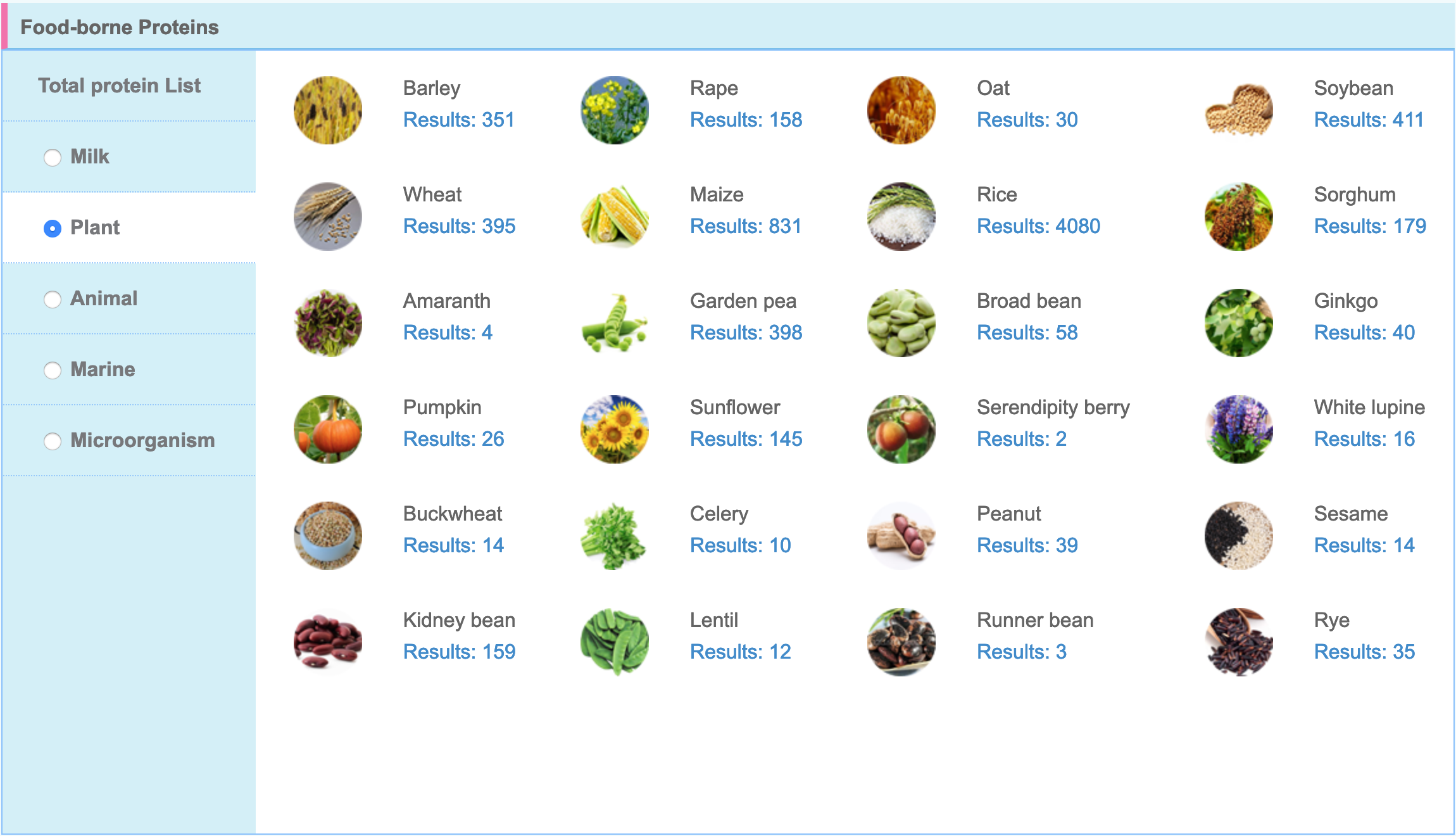
2.2.1 Food-borne protein classification The DFBP database currently contains 21249 food-derived proteins (60 species) reviewed by the Uniprot(https://www.uniprot.org/) database, which are divided into 5 types according to different sources.
|
- Fragments Count: To count the cumulative number of 29 kinds of bioactive peptides in one or a type of protein.
- Frag-Graph: Plot analysis of fragments count data.
- Classification Count: To count the types of 29 kinds of bioactive peptides in one or a type of protein.
- Class-Graph: Plot analysis of classification count data.
|
- Search: To quickly search for proteins by DFBP ID, Organism, Protein name, Protein length and UniProtKB. In order to ensure the convenience of searching, the letter size is not distinguished here.
- Display: The number of proteins displayed on each page can be adjusted by "Display". "First, previous, next, last" is used to quickly turn pages, you can also use "Page to" to directly turn to the specific page.
|
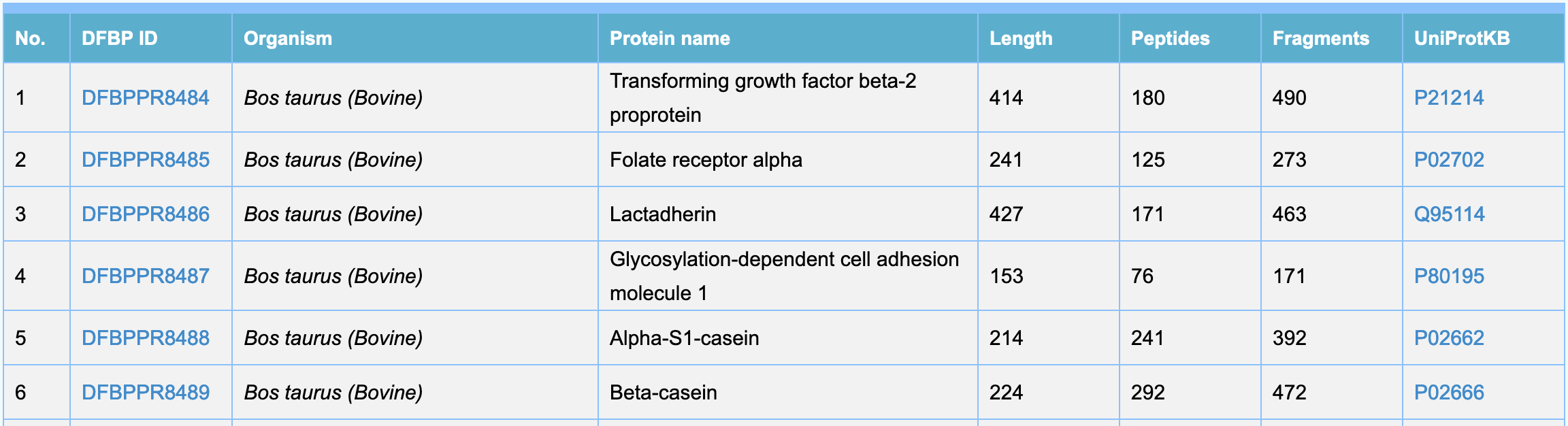
- The brief list is used to display basic information of food-borne proteins, including DFBP ID, Organism, Protein name, Length, Peptides, Fragments and UniProtKB.
- To quickly filter by entering the corresponding information in the quick search tool, results matching the search criteria will be highlighted in the list.
- If you need detailed information of a protein, please click "DFBP ID" number to enter the protein attribute page.
|
2.2.3 Protein attribute page (1) Protein header information
- DFBP ID: The unique ID identification number assigned by DFBP.
- Protein name: Named according to Uniprot (https://www.uniprot.org/).
- Type: All proteins are natural food-borne proteins reviewed by Uniprot.
- Length: The number of amino acid residues.
|
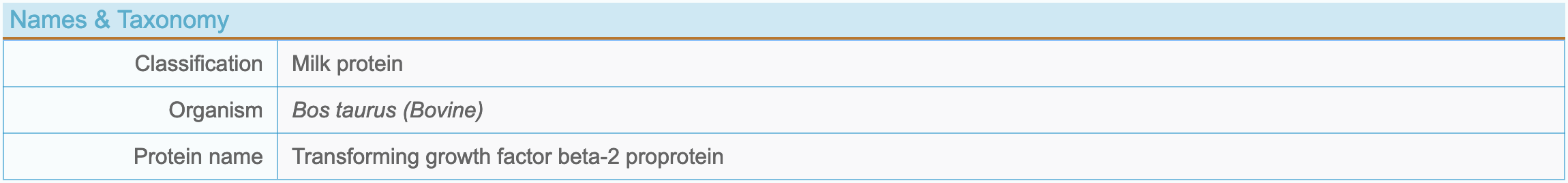
(2) Protein name and taxonomy
- Classification: Food-borne proteins are divided into 5 types based on sources: Milk, Plant, Animal, Marine and Microorganism.
- Organism: To indicate which organism the protein comes from.
- Protein name: Named according to Uniprot (https://www.uniprot.org/).
|
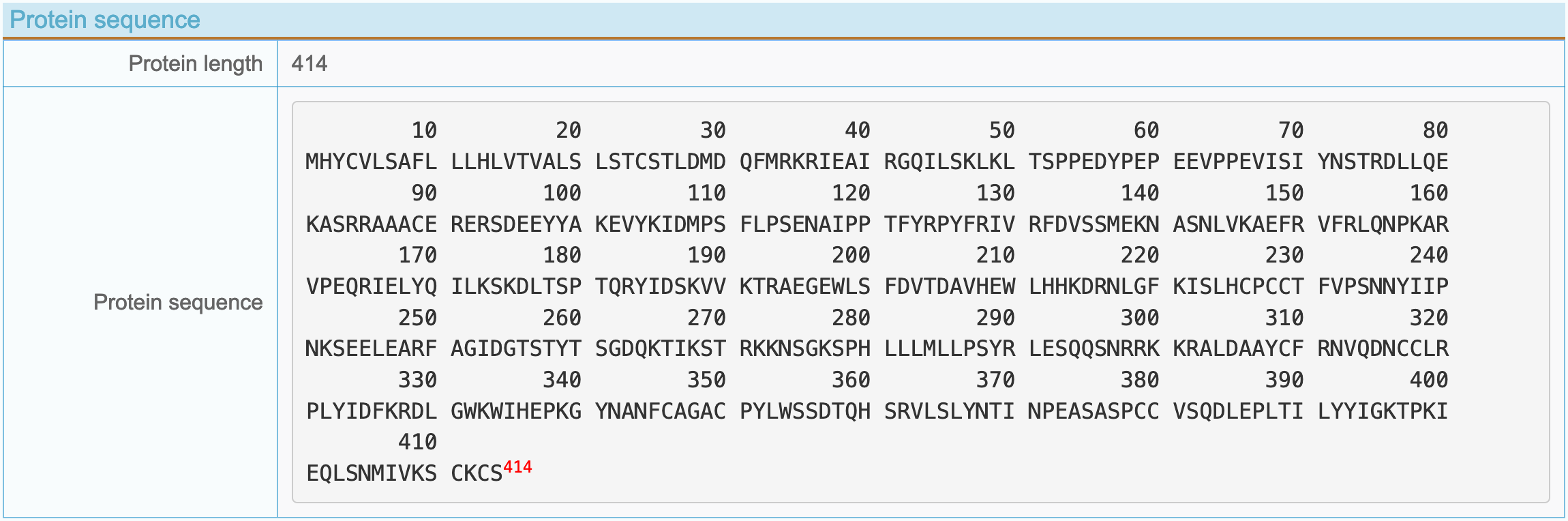
(3) Protein sequence information
- Protein length: The number of amino acid residues in the protein.
- Protein sequence: Single letter amino acid abbreviation sequence.
|
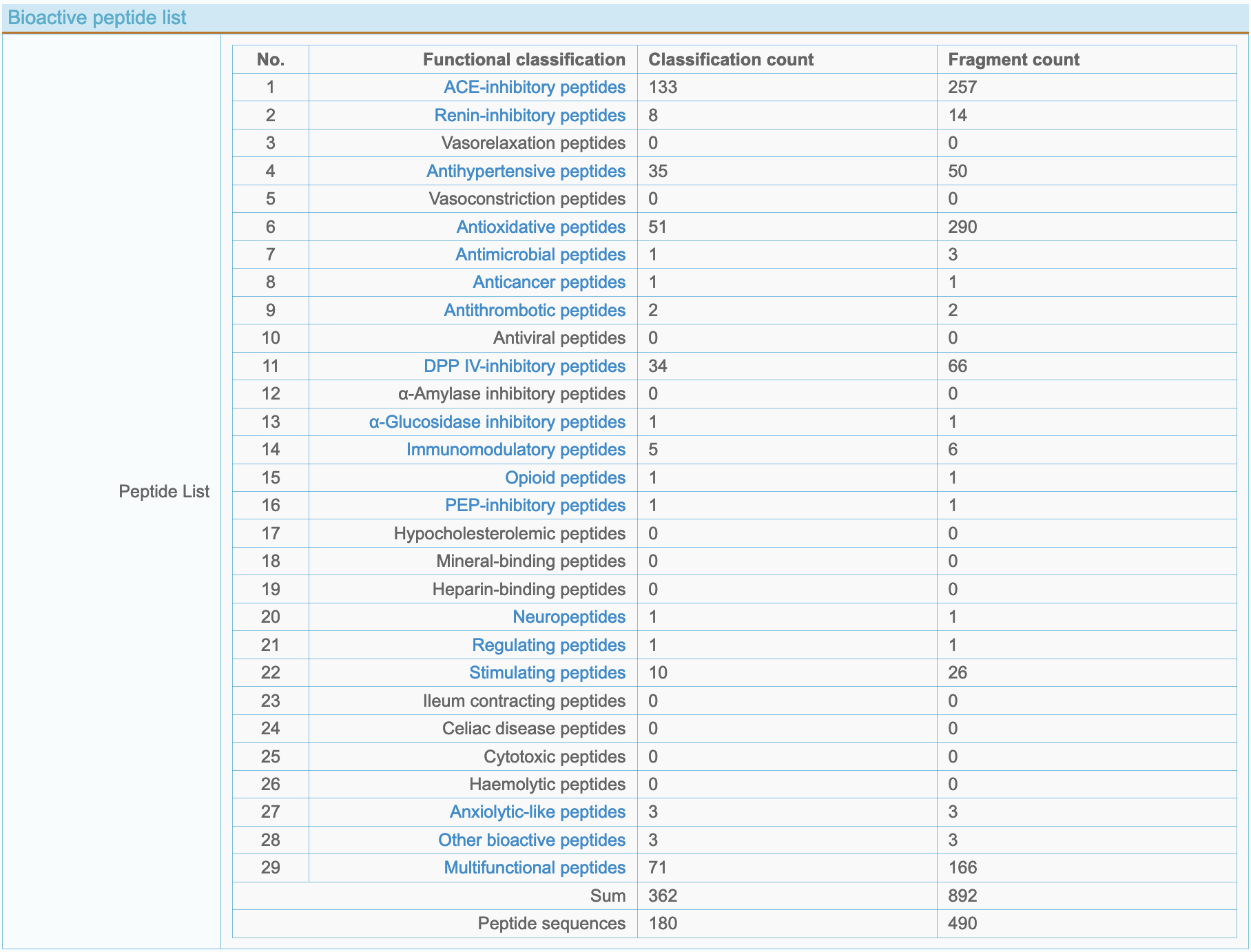
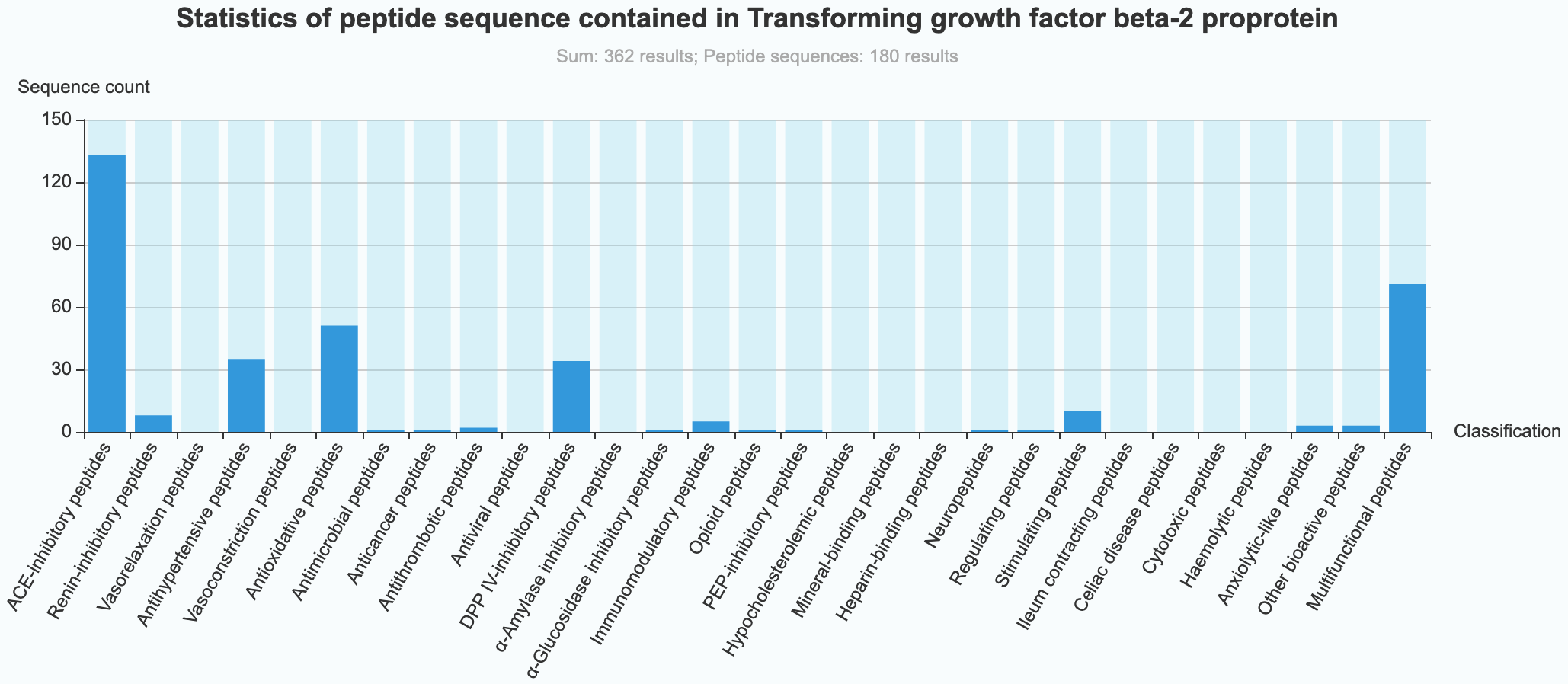
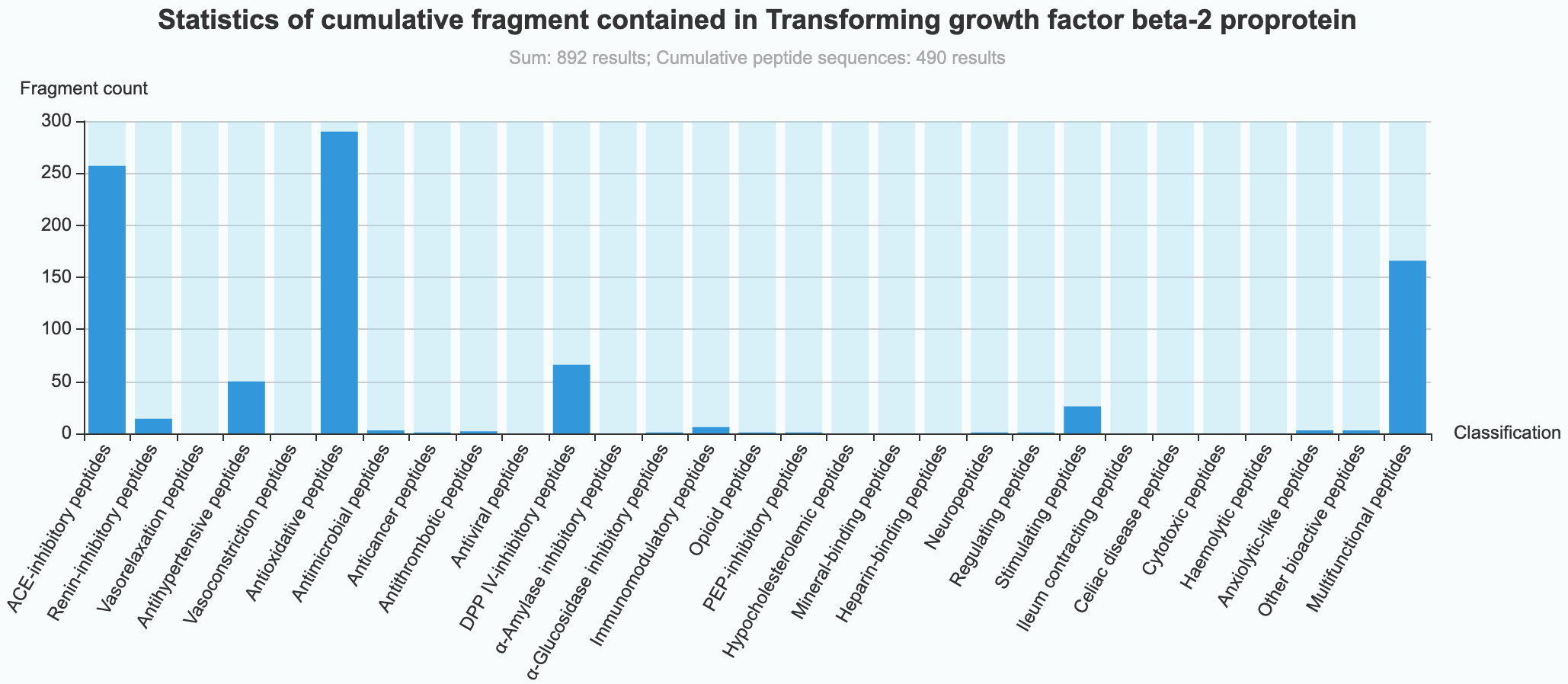
(4) Statistical information of 29 kinds of bioactive peptides in proteins Analyze and count the relevant data of 29 kinds of bioactive peptides in a protein:
- Classification count: Count types of 29 kinds of bioactive peptides in a protein.
- Fragment count: Count the cumulative number of 29 kinds of bioactive peptides in a protein.
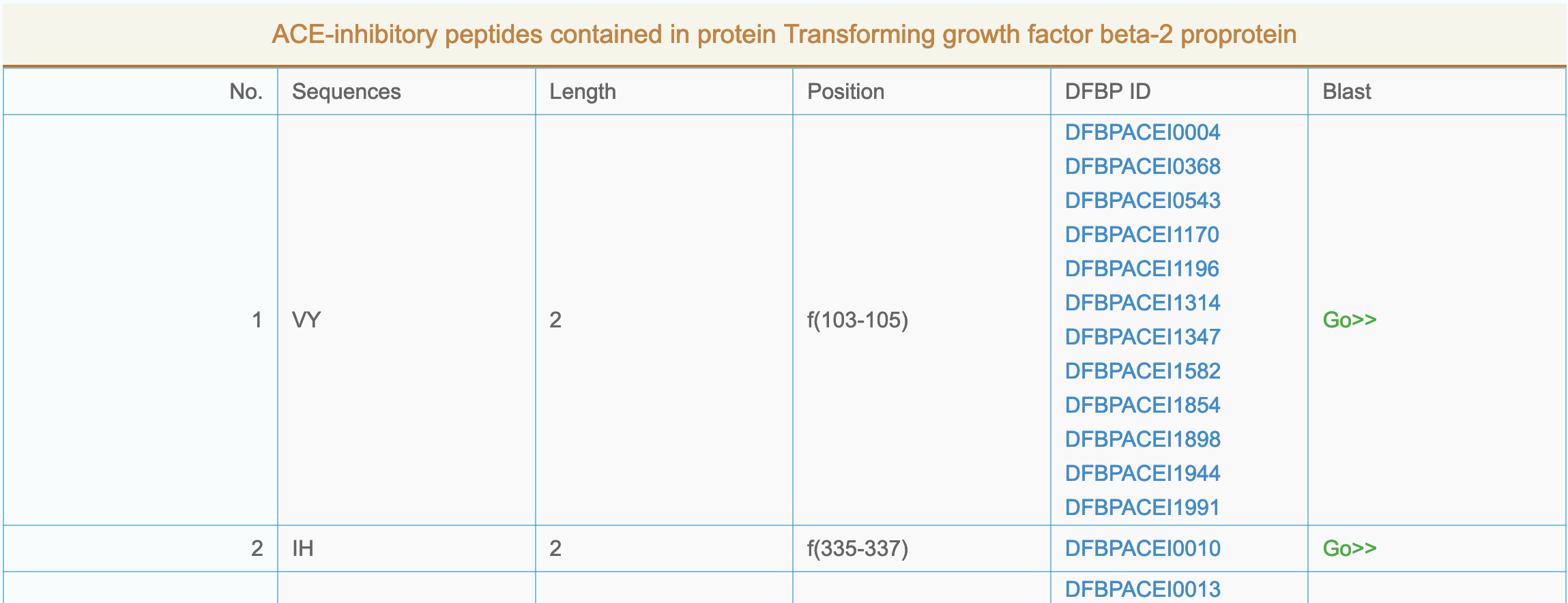
- More info: Click "Functional classification" name to enter the peptide detailed list page, as shown below:
|
(5) Statistical data graph of 29 kinds of bioactive peptides in proteins
- Class-Graph: Plot analysis of types of 29 kinds of bioactive peptides contained in a protein.
- Frag-Graph: Plot analysis of the cumulative data of 29 kinds of bioactive peptides in a protein.
|
(6) Enzymatic hydrolysis prediction
- EHP-Tool: Simulated enzymatic hydrolysis tool provided by DFBP to obtain potential bioactive peptides.
|
(7) Protein cross-references
- Uinprot: Uniprot (https://www.uniprot.org/) reference accession number link.
|